Processed count matrices and metadata, including cell type annotations, can be found here. This Google bucket contains count matrices in the form of anndata object files (.h5ad) for our snRNA and scATAC data. The metadata files (.csv) for barcodes (BCs) and genes are the same as the obs and var data frames, respectively, within the anndata objects. The RNA-seq data has been processed as described in the methods section of the manuscript and detailed in jupyter notebooks prefixed 1-5 on the publication GitHub. Also included in the bucket are count and metadata for all cell types and the GABAergic inhibitory neurons separately. The default count matrix in the RNA anndata objects are the full-raw counts, but the downsampled CPM counts can be found in the anndata layer “ds_norm_cts.” The ATAC anndata object contains processed counts as described in the manuscript. The embeddings presented in the publication are located in the obsm of the anndata objects.
NOTE: Misassigned ‘stage_id’ labels in snRNA-seq BC meta-data corrected as of 18/01/2023, error occurred post-publication while readying data for public consumption.
Interactive Embeddings
| Prefrontal Cortex snRNA-seq | Interneuron snRNA-seq | Prefrontal Cortex snATAC-seq |
|---|---|---|
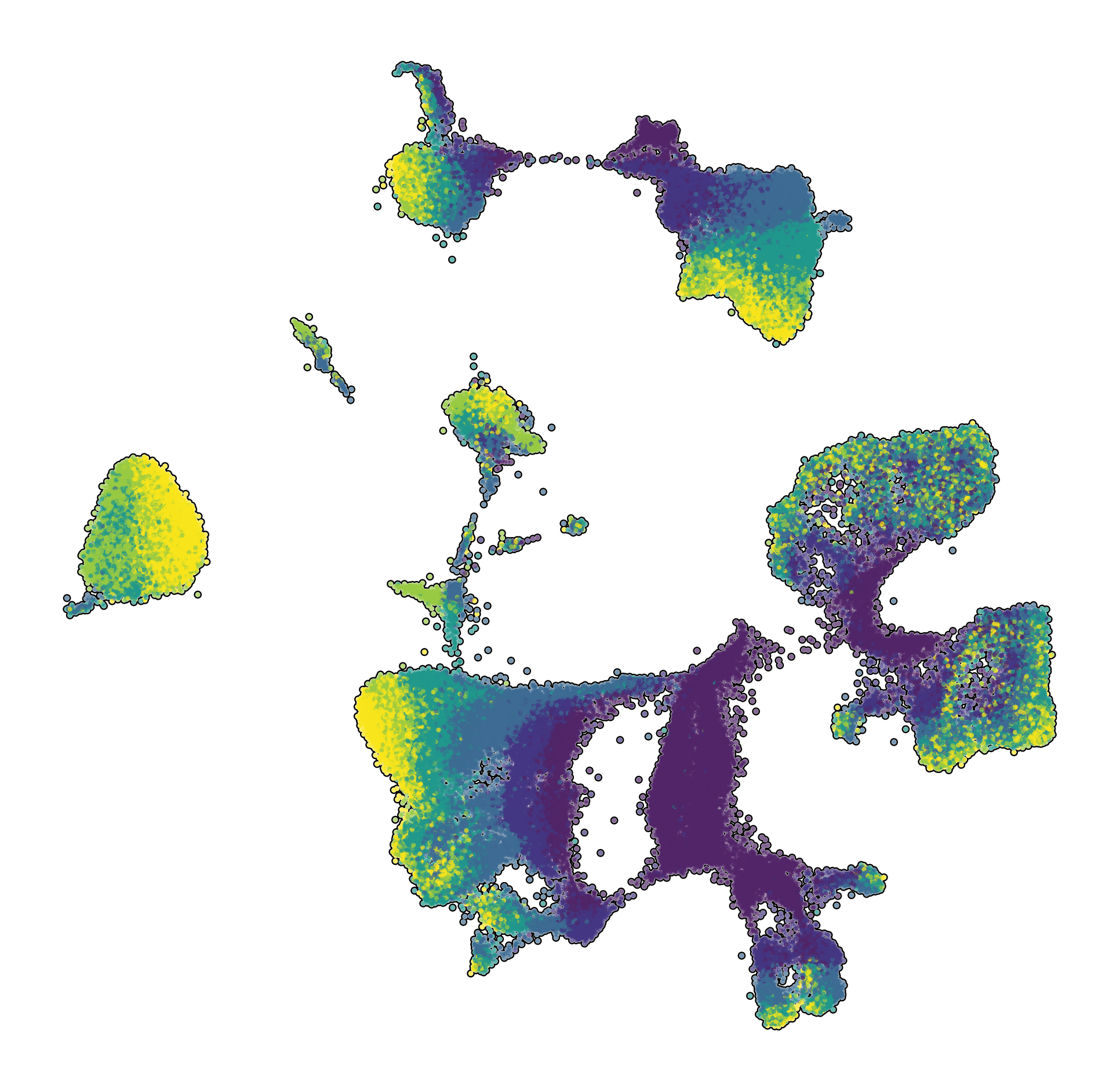 |
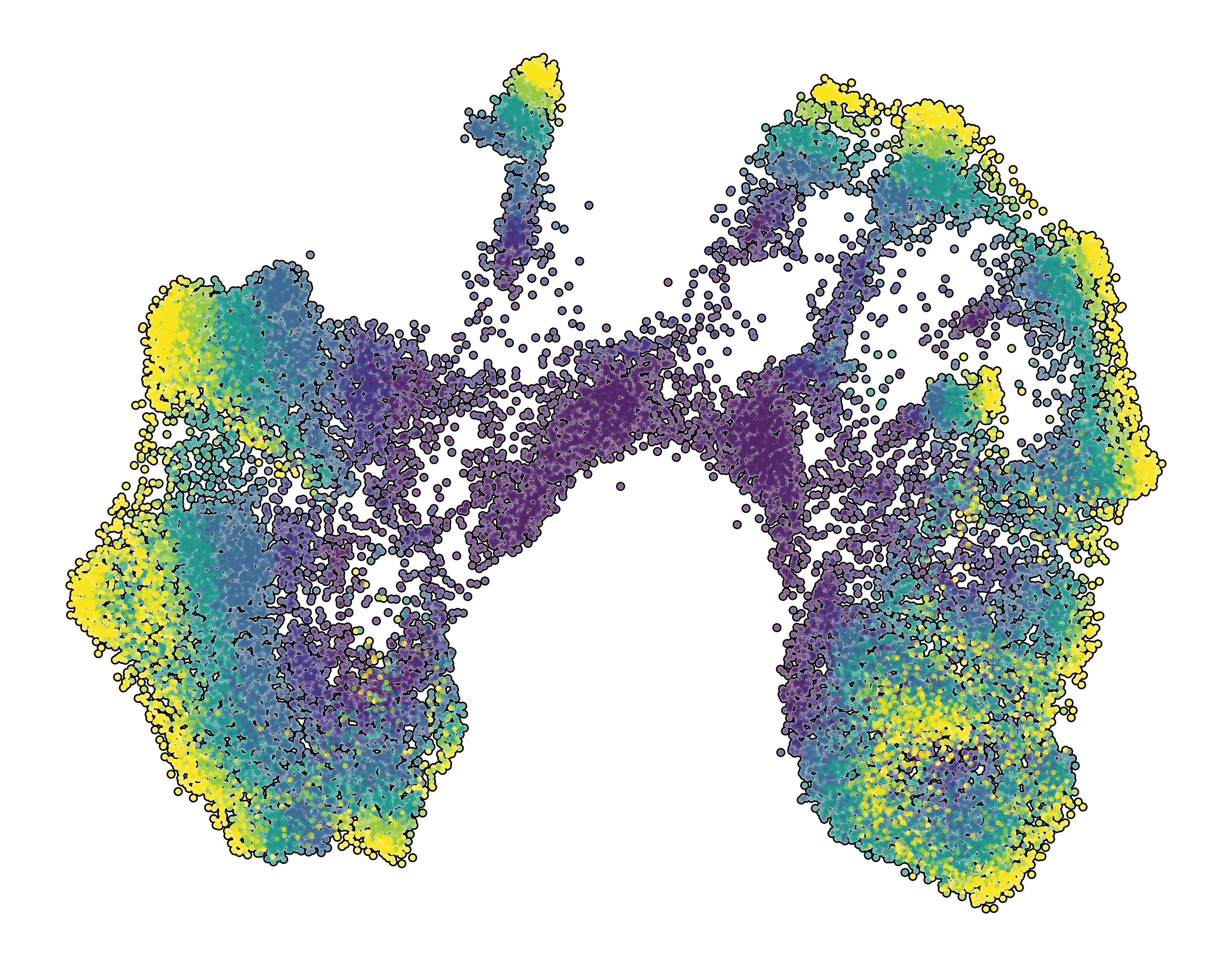 |
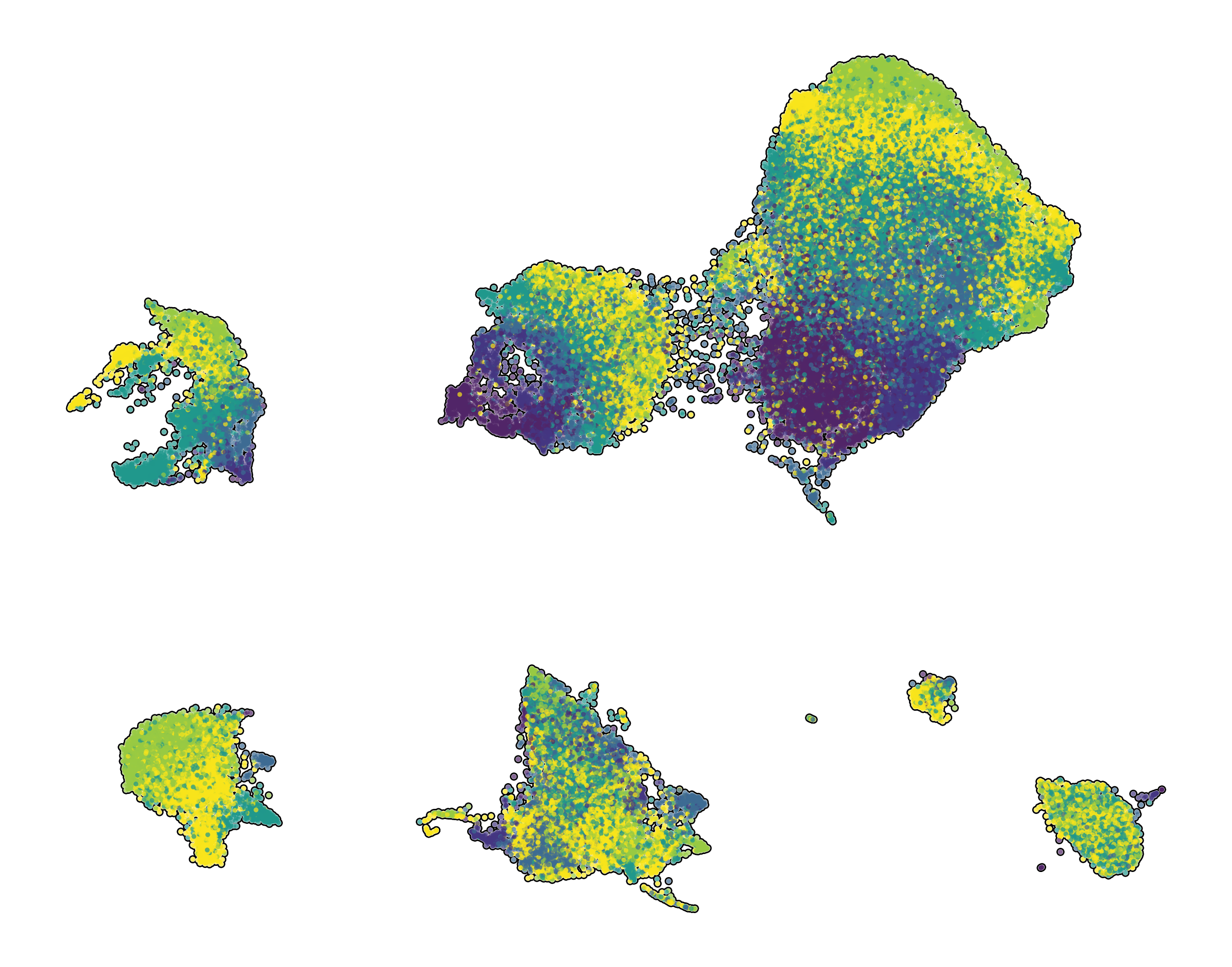 |
Glimma Interactive Graphics for Trajectory devDEGs
| snRNA-seq Major Trajectories | snRNA-seq Sub Trajectories |
|---|---|
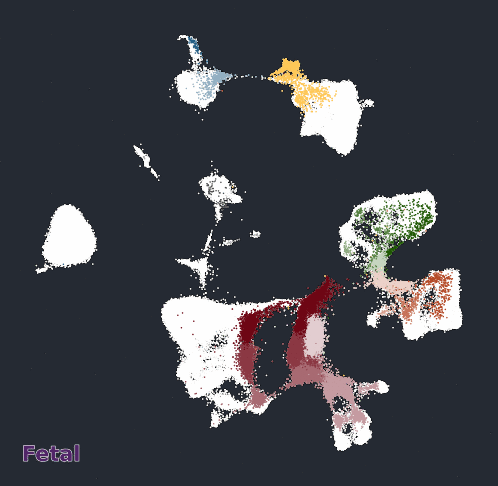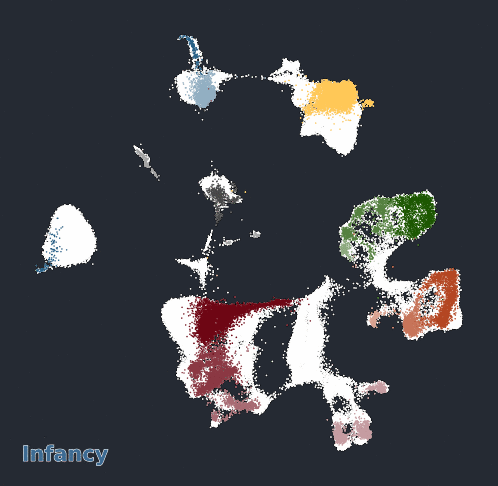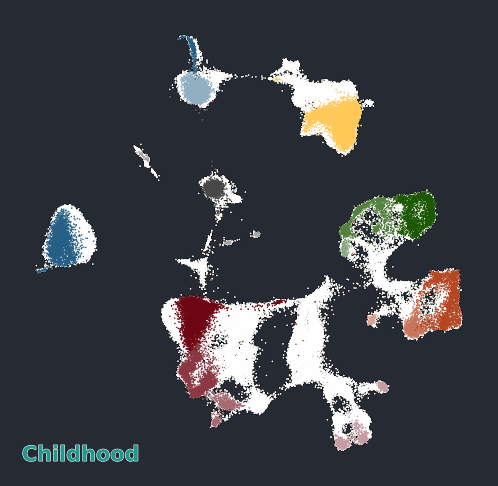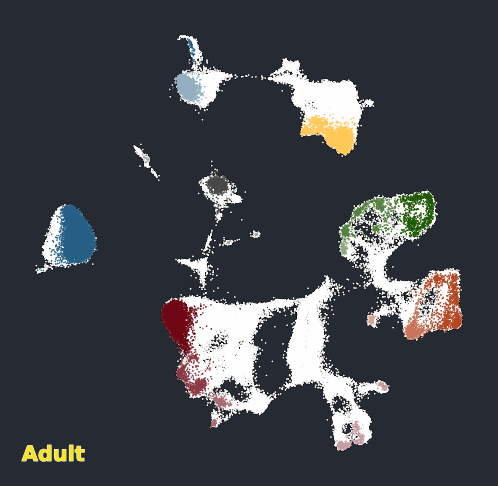 |
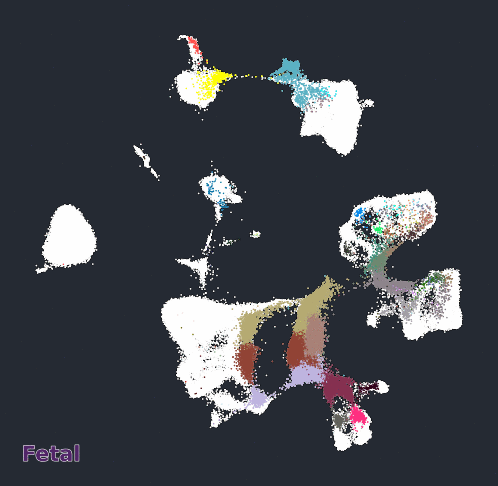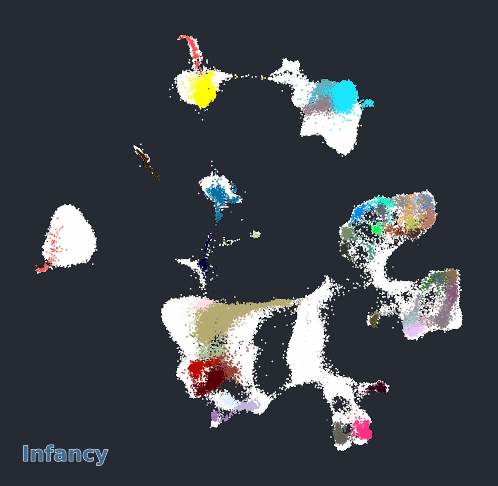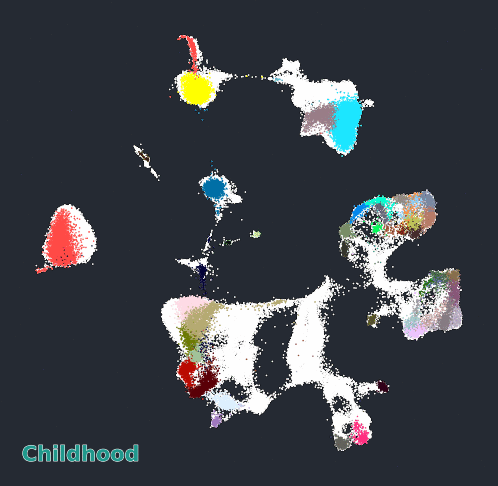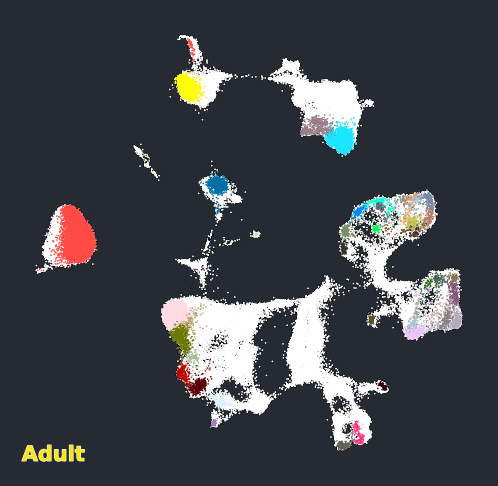 |
Interactive Trajectory devDEG Trend Plots
| snRNA-seq Major Trajectories | snRNA-seq Sub Trajectories |
|---|---|
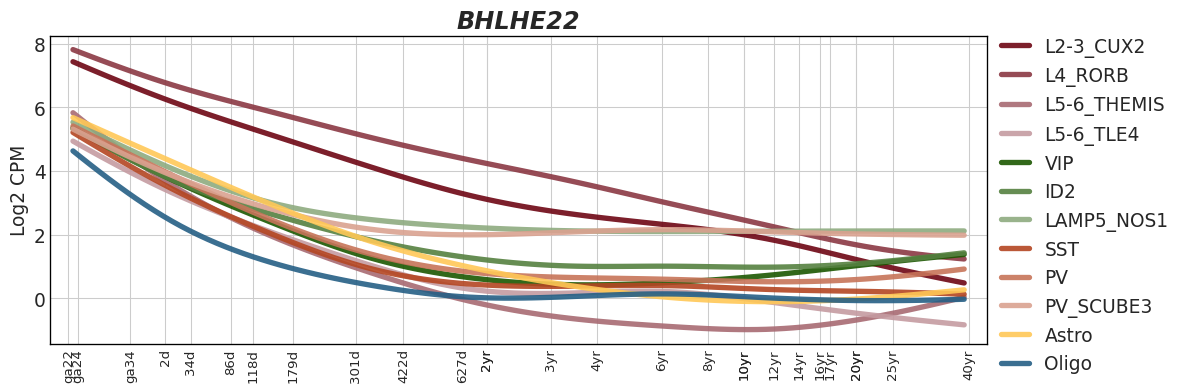 |
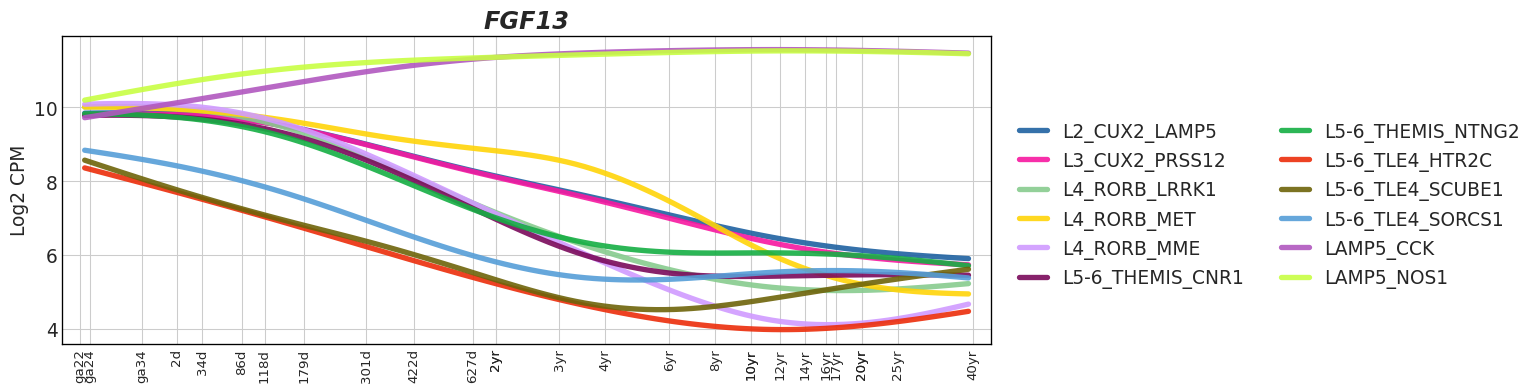 |
Integrative Genomics Viewer
| snATAC-seq IGV Browser Session |
|---|
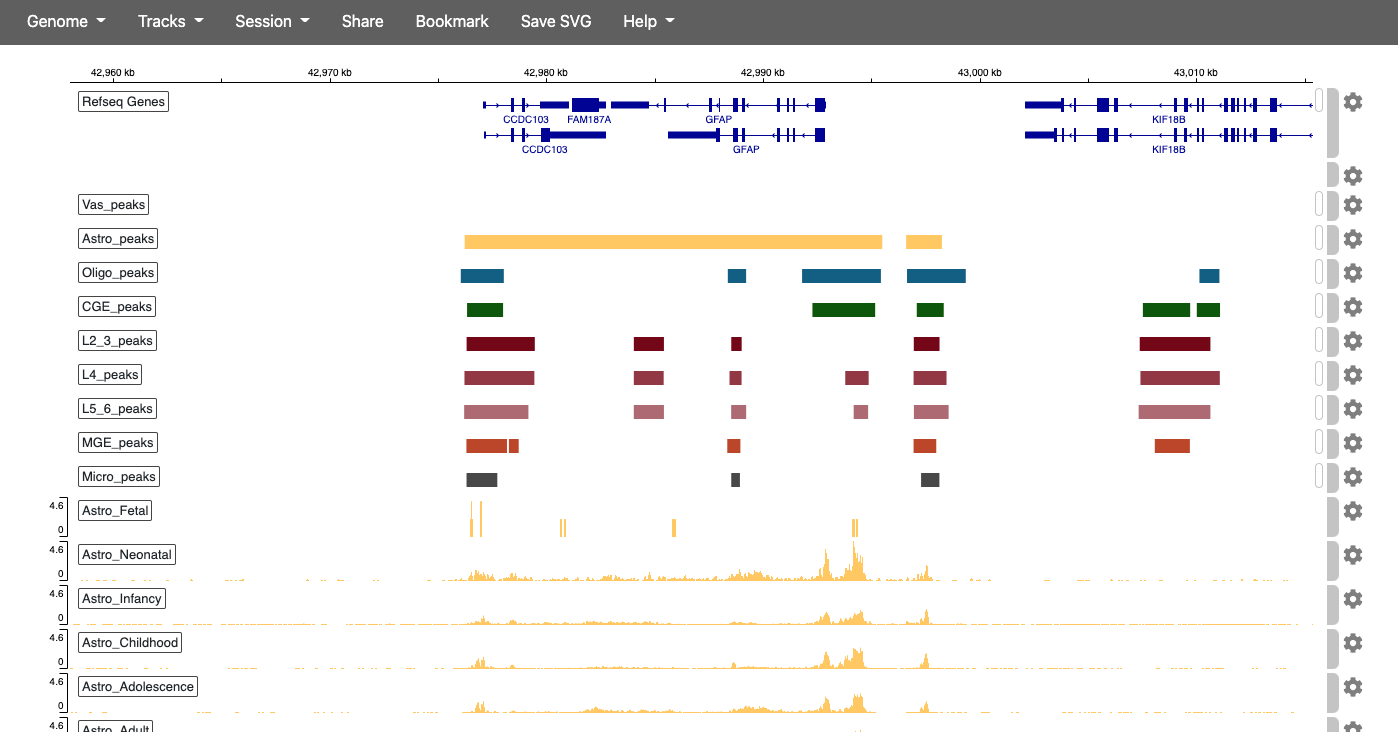 |
